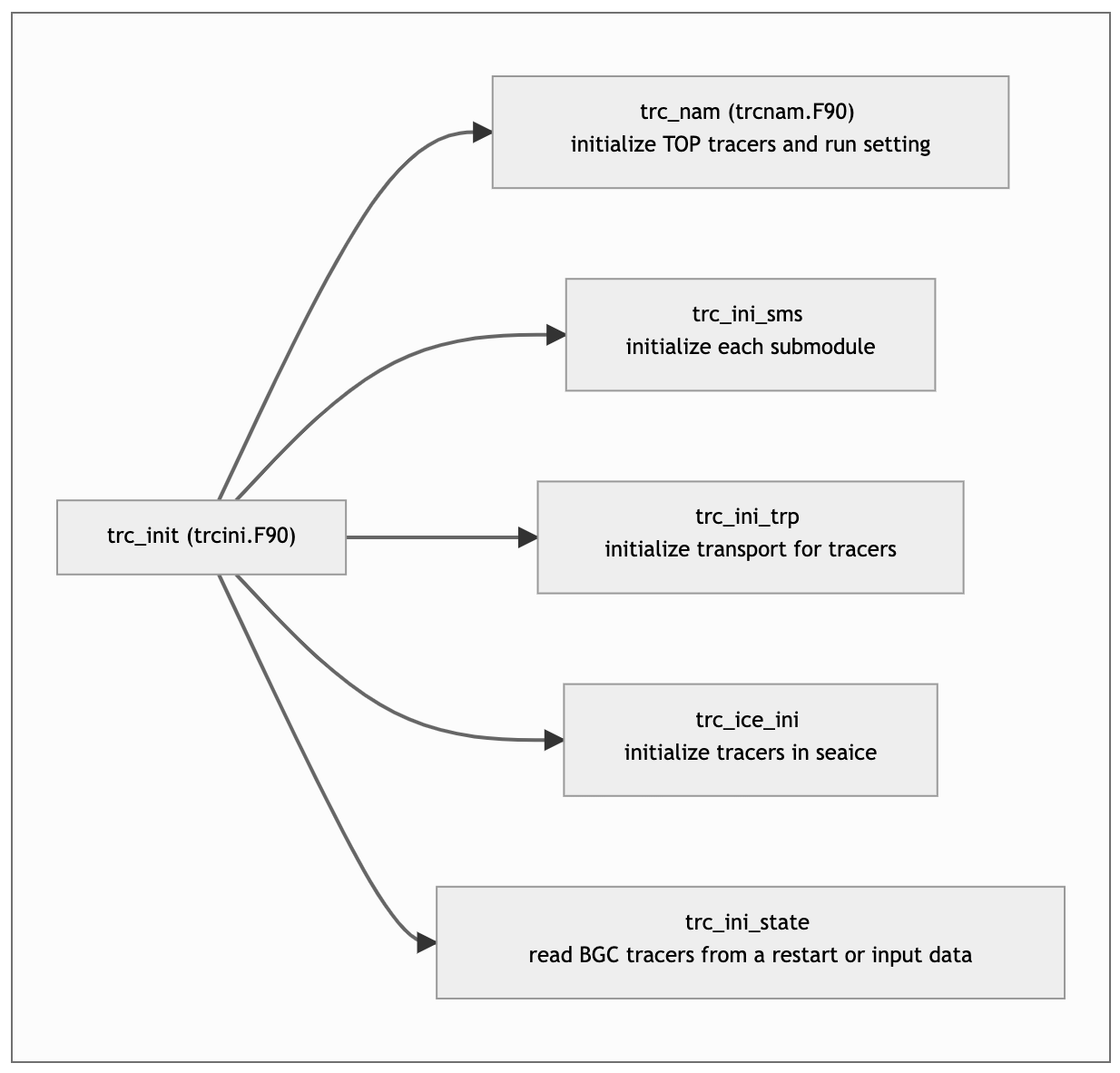
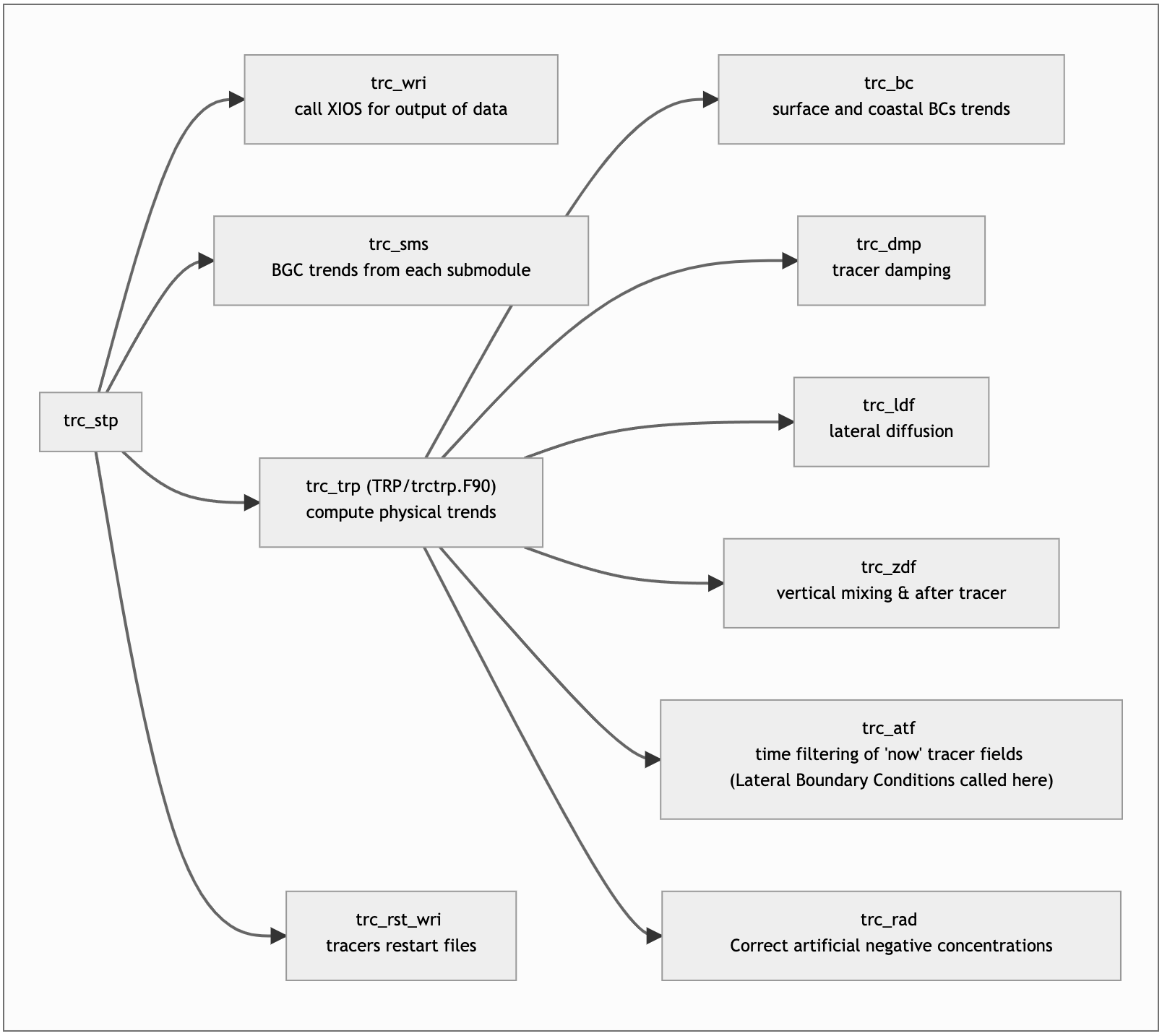
TOP manual structure with figures
Showing
- doc/latex/TOP/figures/TOP_init.png 0 additions, 0 deletionsdoc/latex/TOP/figures/TOP_init.png
- doc/latex/TOP/figures/TOP_step.png 0 additions, 0 deletionsdoc/latex/TOP/figures/TOP_step.png
- doc/latex/TOP/subfiles/model_structure.tex 27 additions, 84 deletionsdoc/latex/TOP/subfiles/model_structure.tex
doc/latex/TOP/figures/TOP_init.png
0 → 100644
112 KiB
doc/latex/TOP/figures/TOP_step.png
0 → 100644
196 KiB